Abstract
Background:
The novel fms-like tyrosine kinase 3 (FLT3)/AXL inhibitor, gilteritinib, showed robust antileukemic activity at doses ≥80 mg/day in FLT3 mutation-positive (FLT3mut+) patients with R/R AML in the CHRYSALIS Phase 1/2 study (NCT02014558). We analyzed variant allele frequency (VAF) in a CHRYSALIS study FLT3mut+ R/R AML subpopulation with baseline FLT3 internal tandem duplication (ITD) and tyrosine kinase domain (TKD) mutations using two different assays: capillary electrophoresis (CE) and next-generation sequencing (NGS). We assessed analytical concordance and survival outcomes at two FLT3 loci: ITD and D835 mutations in the TKD.
Methods:
Assays were performed on bone marrow and blood samples. The CE (LeukoStrat® CDx) FLT3 mutation assay detected both FLT3 -ITD and D835/I836 FLT3 -TKD mutations; these loci were amplified using polymerase chain reaction. Amplicon size differences distinguished samples with FLT3-ITD mutations from those with wild-type FLT3 . Undigested amplicons following restriction enzyme digestion identified the presence of FLT3-TKD mutations. The capture based NGS assay targeted potential mutation loci in all FLT3 exons. Whole genome libraries were generated and hybridized with a custom probe to capture target fragments, which were sequenced on an Illumina® MiSeq platform. The CE FLT3 signal ratio outputs were converted to estimated VAF values for comparison with NGS outputs. For FLT3-ITD analysis, both assays were optimized to detect large ITD mutations. Median overall survival (OS) was determined for patients who received ≥80 mg/day gilteritinib, specifically among those with FLT3-ITD with/without concomitant TKD mutations, and those with TKD mutations only. Median VAF values were calculated; patient VAFs were stratified according to whether they were greater than/equal to, or less than the median value. Lastly, OS of patients with a FLT3-ITD mutation detected by NGS was stratified using a ≥5% VAF cut point.
Results:
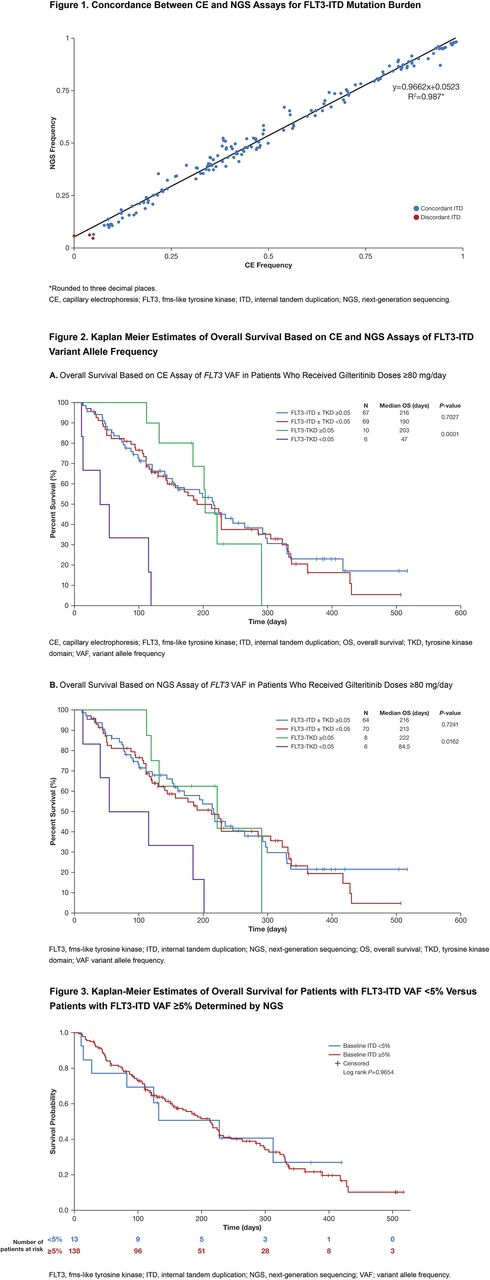
Bone marrow and blood samples from 241 patients were analyzed. The two assays demonstrated strong concordance for both qualitative (mutation detection) and quantitative (VAF) measures at both FLT3-ITD and FLT3-TKD loci. Using a VAF threshold of ≥0.05 and the most prominent ITD in each sample, FLT3-ITD detection by CE or NGS showed a strong concordance in 98.8% (238 of 241) of samples (155 and 156 positives by CE and NGS, respectively). All 3 discordant samples had VAFs that were near (±0.02) the 0.05 VAF threshold and within the 95% CI values for both assays. Strong concordance for FLT3-ITD detection was maintained when multiple ITDs were observed within a sample; a strong linear correlation between both assays was observed for mutation frequencies (R2=0.987) (Figure 1). At a VAF threshold of 0.05, FLT3-TKD mutation concordance was observed in 98.3% (236/240) of samples (34 and 30 positives by CE and NGS, respectively). The 4 discordant samples were near (±0.04) the 0.05 VAF threshold and there was a strong a linear correlation between observed mutation frequencies (R2=0.906).
At a VAF threshold of ≥5% for FLT3 mutation positivity, there was no apparent difference in OS in the ≥80 mg/day gilteritinib subgroup among those with FLT3-ITD with/without concomitant TKD mutations who had VAF values that were greater than/equal to or less than the median values (Figure 2A and 2B), regardless of the assay used. Patients who had a TKD mutation only with VAF values greater than/equal to the median value had longer OS than patients with VAF values less than the median value. Patients who had a TKD mutation only with VAF values greater than/equal to the median value had OS durations similar to those in patients with ITD mutations. By NGS, patients with a FLT3 -ITD VAF <5% had similar OS duration as patients with a FLT 3-ITD VAF ≥5% (Figure 3).
Conclusions:
We observed high concordance using both CE and NGS methods to measure FLT3 mutational burden in patients with R/R AML who received gilteritinib in the CHRYSALIS study. Analytical concordance between the 2 assays was high and survival outcomes with stratification based on FLT3 mutational burden were similar with both assays. Our results suggest that FLT3-ITDmut+ R/R AML patients may benefit from gilteritinib therapy, regardless of FLT3 mutational burden. In patients with FLT3-TKD mutations only, a higher mutational burden may be associated with gilteritinib benefit; this finding will be further explored in future studies.
Smith: Plexxikon Inc.: Research Funding; Astellas Pharma: Research Funding. Perl: Pfizer: Other: Advisory Board; Actinium Pharmaceuticals: Other: Scientific Advisory Board; Asana Biosciences: Other: Scientific advisory board; Astellas: Consultancy; Novartis: Other: Advisory Board; Seattle Genetics: Other: Advisory board; Daiichi Sankyo: Consultancy; Arog Pharmaceuticals: Consultancy. Altman: Syros: Consultancy; NCCN: Other: Educational speaker; BMS: Consultancy; Celgene: Consultancy; Astellas: Consultancy; Ceplene: Consultancy; Janssen Pharmaceuticals: Consultancy; Novartis: Consultancy; ASH: Other: Educational speaker. Carson: Invivoscribe, Inc.: Employment. Cortes: ImmunoGen: Consultancy, Research Funding; Teva: Research Funding; Novartis Pharmaceuticals Corporation: Consultancy, Research Funding; ARIAD: Consultancy, Research Funding; Sun Pharma: Research Funding; Pfizer: Consultancy, Research Funding; BMS: Consultancy, Research Funding. Stenzel: Invivoscribe, Inc.: Employment. Hill: Ligacept, LLC: Equity Ownership; Patent: Patents & Royalties: WO2013163419A1 - pending; Patent: Patents & Royalties: US7862995B2 - Issued; Astellas Global Pharma Development: Employment. Lu: Astellas Pharma Global Development: Employment. Bahceci: Astellas Pharma Global Development: Employment. Levis: Daiichi Sankyo, Inc.: Consultancy, Honoraria; FujiFilm: Research Funding; Astellas Pharma Us: Consultancy, Research Funding; Millennium Pharmaceuticals, Inc., a wholly owned subsidiary of Takeda Pharmaceutical Company Limited: Research Funding; Novartis: Consultancy, Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal